Importing Data
Importing data via buttons
The data import features can be accessed from the environment pane or from the tools menu. The importers are grouped into 3 categories: Text data, Excel data and statistical data. To access this feature, use the “Import Dataset” dropdown from the “Environment” pane:
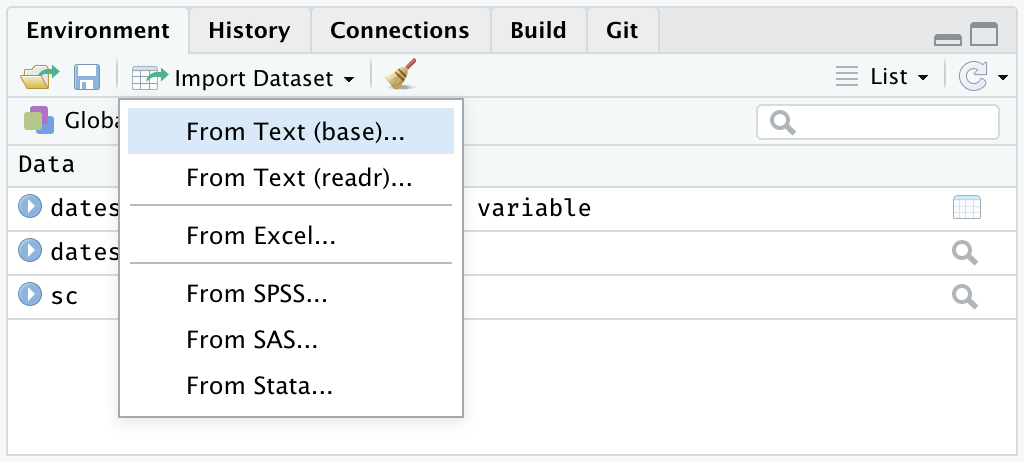
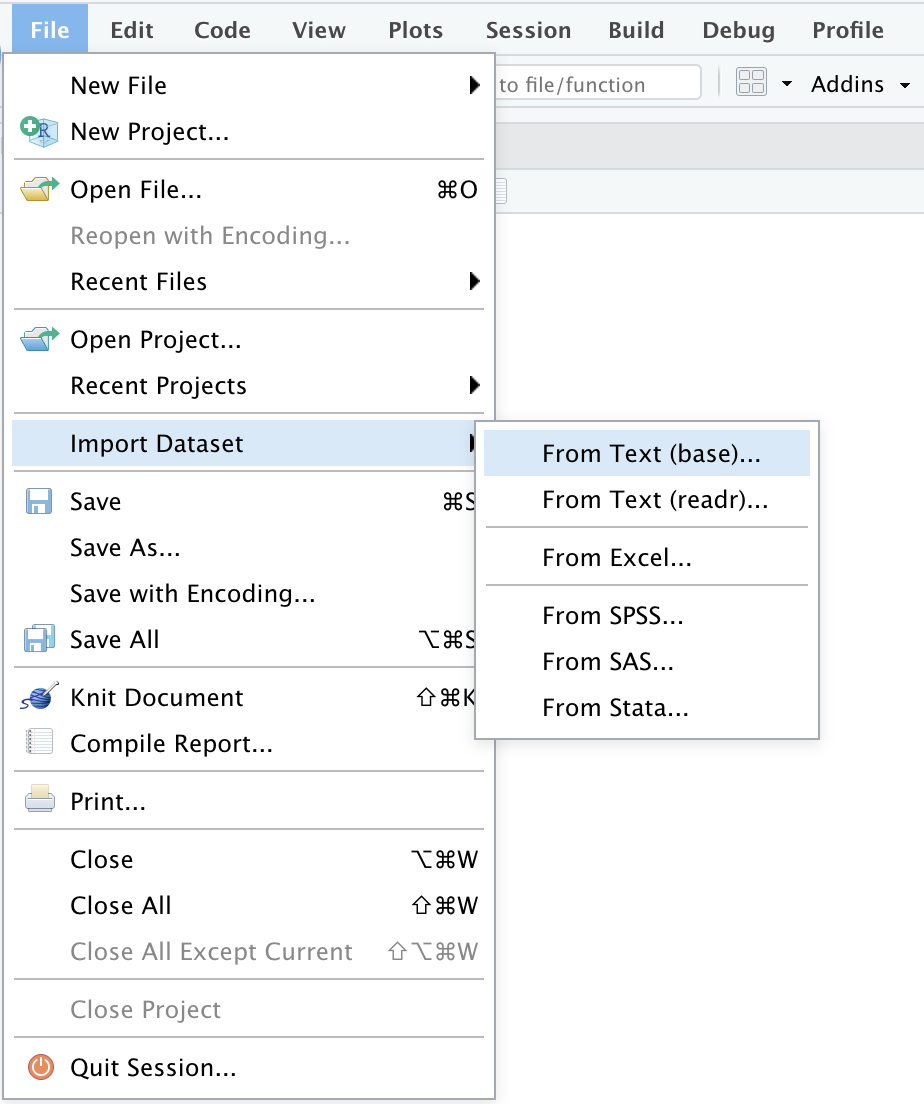
Or through the “File” menu, followed by the “Import Dataset” submenu:
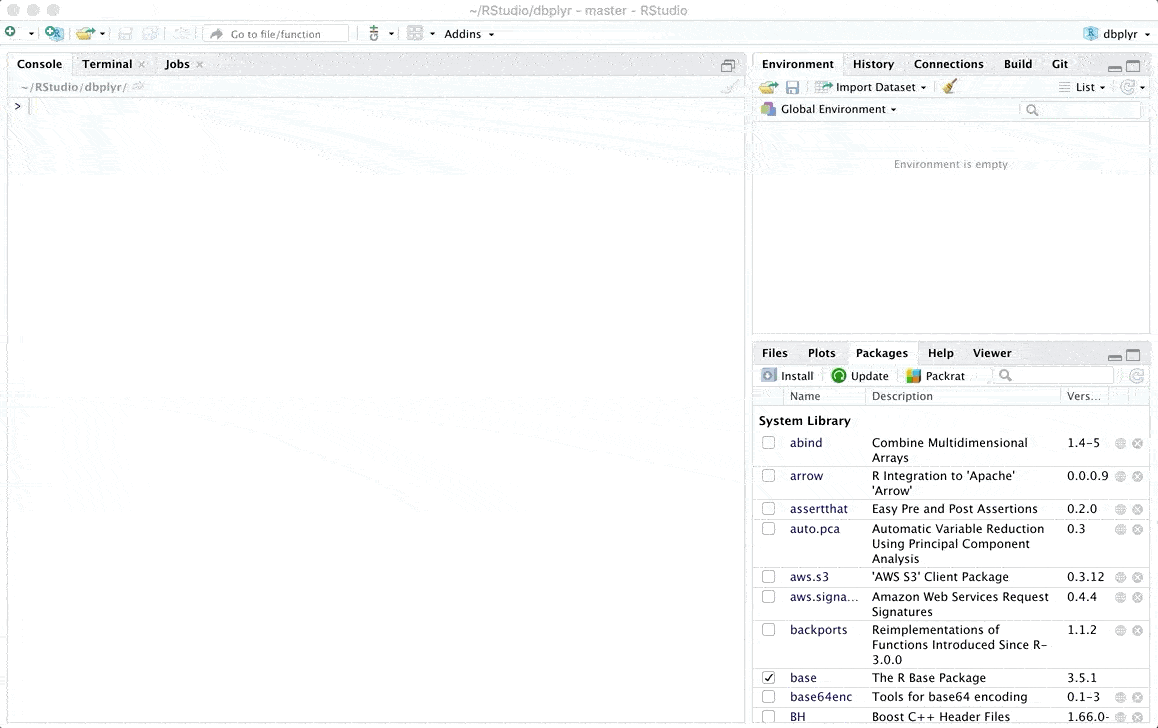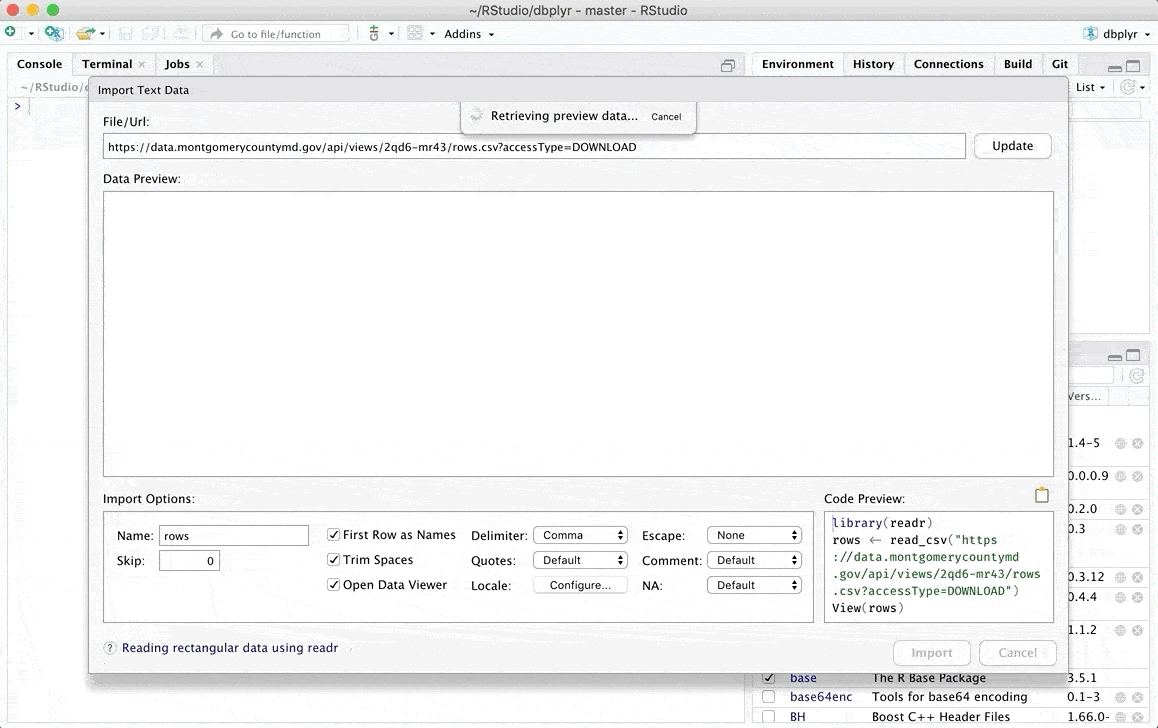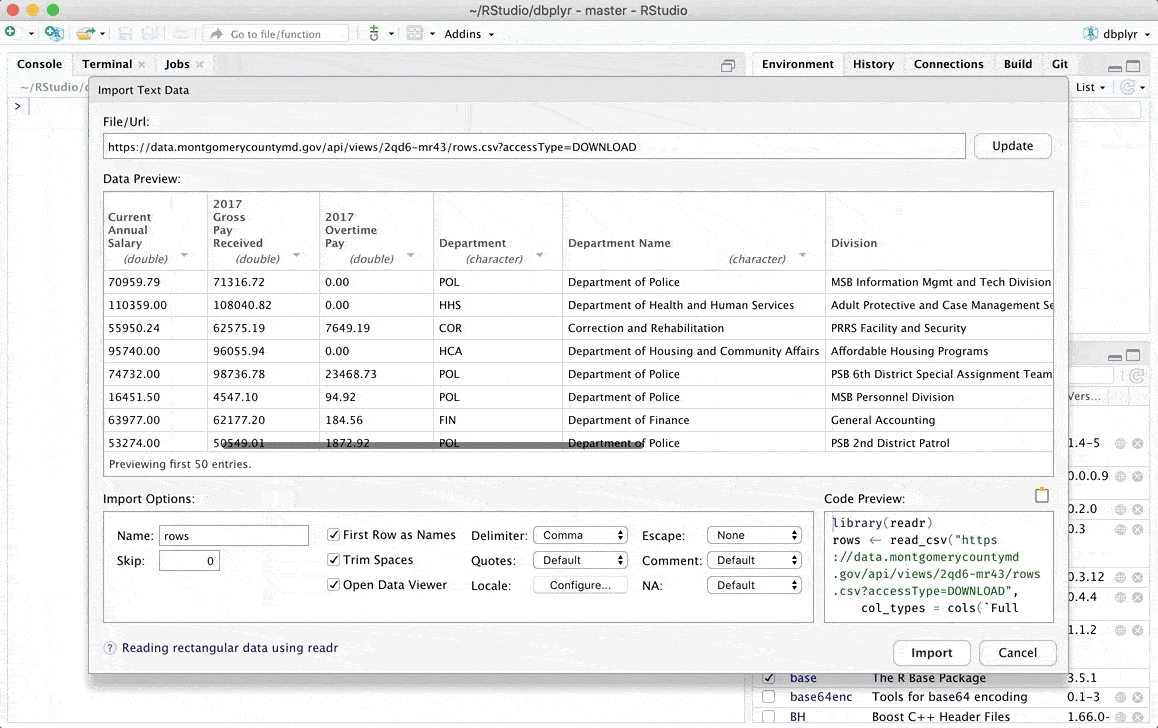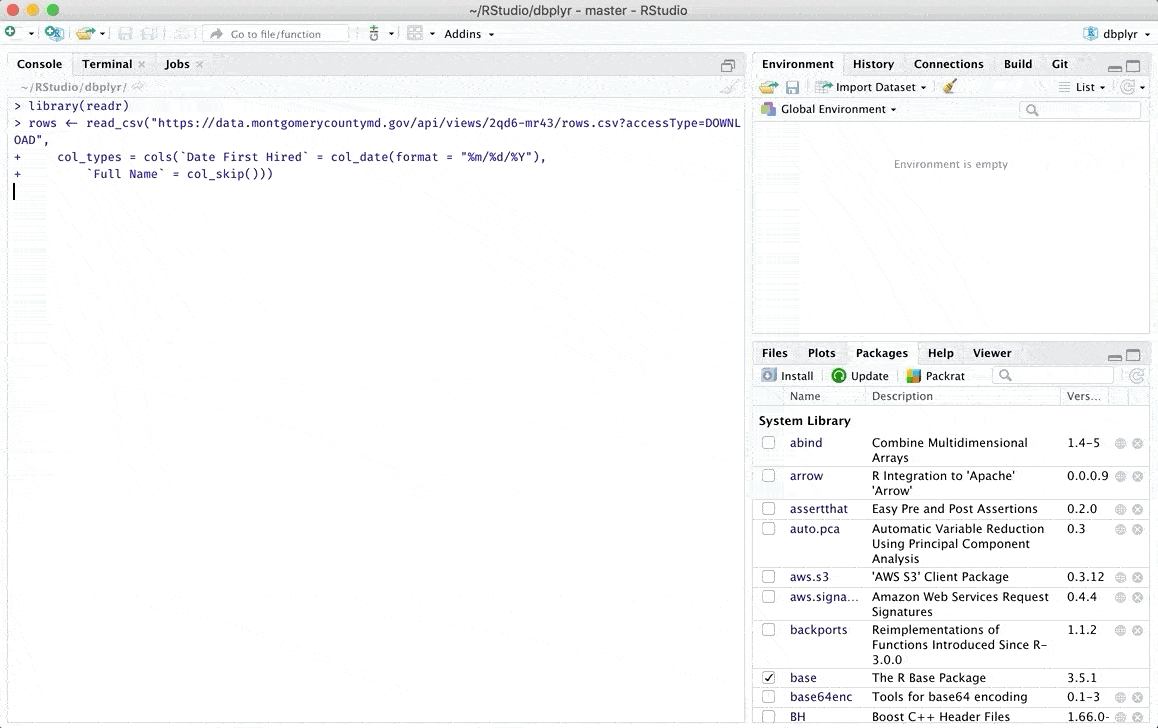
Importing From Text (readr) files allows you to import csv files and in general, character delimited files using the readr package. This Text importer provides support to:
- Import from the file system or an url
- Change column data types
- Skip or include-only columns
- Rename the data set
- Skip the first n rows
- Use the header row for column names
- Trim spaces in names
- Change the column delimiter
- Encoding selection
- Select quote, escape, comment and NA identifiers
Importing data via code
Reading or writing tabulated data into or from a data frame is a quite common task in data analysis. The read.table or the read.csv functions for this.
df <- read.csv(file.csv) # or read.csv2 -> selection depends on the delimiter and seperator
df <- read.table(file.csv, header = FALSE, sep = “”,
dec = “.”, skip = 0, ...)
write.table(df, file = <file>, sep = “ “, dec = “.”, ...)
Reading data from csv files
Reading csv files is realized using the read.table function from R’s utils library. The function will return a data frame which contains the information of the csv file (example taken from here) .
df <- read.table(paste0(envrmt$path_data_csv, "/AI001_gebiet_flaeche.csv"),
skip = 4, header = TRUE, sep = ";", dec = ",")
As you can see, the read.table function gets several arguments (which is common for many functions). The first one gives the filename inclducing the path to the file.
skip = 4tells the function to skip the first four lines (which are plain text lines in this case and not tabulated values)header = TRUEtells the function, that the csv file has a header line which is used byread.tableto name the columns of the returning data framesep = ";"defines the separator of the individual columns in the data framedec = ","defines the decimal separator used in the dataset
A note on the sequence of the arguments: the sequence of the arguments does
not matter as long as you name them explicetly. If you do not use the argument
identfier as it is the case for the first argument, the filename, in the example
then the sequence matters. To get information on the default sequence and of
course the general application of the each R function, type ?<function name>
(e.g. ?read.table) in an R console.
After you executing the read.table function above, the content of the csv file is
stored into a two dimensional data frame called df in the example above.
A quick way to check if everything is fine is to display the first few lines of
the data file using the head function (without the 2, it will print 5 lines as a standard setting).
head(df,2)
## X X.1 X.2
## 1 1996 DG Deutschland
## 2 1996 01 Schleswig-Holstein
## Anteil.Siedlungs..und.Verkehrsfläche.an.Gesamtfl.
## 1 11,8
## 2 10,8
## Anteil.Erholungsfläche.an.Gesamtfläche
## 1 0,7
## 2 0,7
## Anteil.Landwirtschaftsfläche.an.Gesamtfläche
## 1 54,1
## 2 73,0
## Anteil.Waldfläche.an.Gesamtfläche
## 1 29,4
## 2 9,3
Writing data to csv files
Writing data is as easy as reading it. Just use the write.table function.
write.table(df, file = paste0(envrmt$path_data_tmp, "new.csv"),
sep = ",", dec = ".")
As you can see, you can define any column or decimal separator.
For more information have a look at e.g. the respective data importing and data exporting site at Quick R, have a look into the package documentation or search the web.
Alternative data I/O using RDS
Writing into CSV files is a good choice for data exchange into the non-R world. If you want to re-use the information stored in a data.frame or any other variable in R, writing the actual R object to a file connection will be of some benefit especially for more complex objects like model outputs or geo-datasets which do not represent the final information (because this would likely be shared to others using GeoTiff or other well recognized formats). To save an R object to a file connection, use the saveRDS function, to read it, use the readRDS function. The file extension .rds is generally used for that format.
If you stay within R for reading and writing R objects from and to data files, you could also use the serialization of readRDS and saveRDS.
saveRDS(df, file = paste0(envrmt$path_data_tmp, "new.rds"))
# Read data to different variable
df2 = readRDS(paste0(envrmt$path_data_tmp, "new.rds"))
saveRDS(df, file = <file>)
df = readRDS(<file>)
See the respective help pages for more details.
Reading or writing tabulated data into or from a data frame is a quite common task in data analysis. You could use the read.table function for this.
df <- read.table(<file>, header = FALSE, sep = “”,
dec = “.”, skip = 0, ...)
write.table(df, file = <file>, sep = “ “, dec = “.”, ...)
Writing into CSV files is a good choice for data exchange into the non-R world. If you want to re-use the information stored in a data.frame or any other variable in R, writing the actual R object to a file connection will be of some benefit especially for more complex objects like model outputs or geo-datasets which do not represent the final information (because this would likely be shared to others using GeoTiff or other well recognized formats). To save an R object to a file connection, use the saveRDS function, to read it, use the readRDS function. The file extension “rds” is generally used for that format.
saveRDS(df, file = <file>)
df = readRDS(<file>)
See the respective help pages for more details.
