| EX | A randomly good model |
Create a random forest model with random cross-validation.
Set up a working environment
First, set up the working environment as described in Unit 01. Add some more packages and additional folders to your setup script. In the “modelling” folder you can store your prepared data, your models, your predictions etc.
# setup call using the default values for the course
require(envimaR)
packagesToLoad = c("mapview", "raster", "sf", "caret", "exactextractr",
"doParallel", "CAST", "ranger")
projectDirList = c("data/",
"data/modelling/",
"data/modelling/model_training_data/",
"data/modelling/models/",
"data/modelling/prediction/",
"data/modelling/validation/",
"docs/",
"run/",
"tmp",
"src/",
"src/functions/")
Read the data
In the first step, you need to import your previously prepared predictor variables (DOP and indices) as well as the building polygons. Combine all of your raster layers into one raster stack. Finally, make sure that each layer has a unique name.
# load rasterStack containing red, green, blue and NIR bands and some indices
rasterStack = readRDS(file.path(envrmt$path_data, "dop_indices.RDS"))
# check that all names are unique
names(rasterStack)
At this point, it is also a good idea to check that the raster and polygons have the same Coordinate Reference System (CRS). A CRS is a set of parameters that determine how to display geographic coordinates (i.e. tell your computer how to represent the Earth and your data on your screen). Also, we need to make sure that each polygon has a unique ID. If the data does not include an ID number, add one.
# now the vector data: use the sf package to load the file and check if the crs matches with the raster stack
# use the vector data you created with QGIS here:
pol = sf::read_sf(file.path(envrmt$path_data,"training_data.gpkg"))
pol = sf::st_transform(pol, crs(rasterStack))
# add IDs to your polygons, if they don't have them
pol$OBJ_ID = 1:nrow(pol)
# if you have more than two classes combine them
pol[pol$class != "buildings",]$class <- "other"
Extract the data
Next, we want to extract the values from the predictor raster stack for every pixel in the training polygons. You can think about the polygons like cookie cutters and the raster stack like several layers of cookie dough stacked on top of each other – we want to cut the data out of each raster that corresponds to the polygon area, i.e. the buildings.
The data gets formatted in a dataframe that we can merge with the original polygons, so that it returns the class information of the polygons.
In the previous Unit we used the extract function of the raster package as it is easy to use.
A somewhat faster approach is to use the extract function of the newer terra package, but it is still quite slow.
As we have a lot of pixels due to the high resolution of the DOP we will use a specific package just for raster extraction here.
The package exactextractr provides the function exact_extract, which we will use here, as it enables us to extract the data faster.
# to extract the data use raster::extract or the much faster package exactextractr
extr = exact_extract(rasterStack, pol, include_cols = c( "class", "OBJ_ID"))
extr = extr[sapply(extr, is.data.frame)]
extr = do.call(rbind, extr)
saveRDS(extr, file.path(envrmt$path_model_training_data, "extraction.RDS"))
Balancing
In some cases, you may want to use machine learning algorithms on data that is imbalanced. This occurs when the number of training polygons for one class is significantly larger (or smaller) than that of the other classes in the dataset. If we used imbalanced data, we may create a model that cannot properly predict for unknown data, because one class is either over- or underrepresented in the training data.
In our next step, we balance the data to counteract this potential error. To do so, we will use the dataframe with the extracted values for each pixel. First, we need to check how many pixel of each class are in our dataframe. Then, we downsample the larger class. This entails taking a random sample of rows from the larger class that is equal to the number of rows in the smaller class.
extr = readRDS(file.path(envrmt$path_model_training_data, "extraction.RDS"))
extr = na.omit(extr)
buildings = extr[extr$class == "buildings",]
other = extr[extr$class == "other",]
nrow(buildings)
nrow(other)
# reduce the larger class by sampling the same number of rows that the smaller class has
set.seed(42)
other = other[sample(nrow(buildings)), ]
# combine the two dataframes
extr = rbind(other, buildings)
Then, we split the data into two sets: one for model training and the other for testing. Here, we use 80% of the data for training and 20% for testing. We will use the function createDataPartition from the caret package for this task. This function is very useful, as it tries to maintain the ratio of each class in the datasets.
trainIndex = caret::createDataPartition(extr$class, p = 0.8, list = FALSE)
training = extr[ trainIndex,]
testing = extr[ -trainIndex,]
saveRDS(training, file.path(envrmt$path_model_training_data, "extr_train.RDS"))
saveRDS(testing, file.path(envrmt$path_model_training_data, "extr_test.RDS"))
Random forest
Now we can finally start our first attempt at predicting the buildings. As discussed before, we will use a simple random forest model with 10-fold cross-validation. Define your train control settings and use the train function from caret (on CRAN) to train your model. For an in-depth understanding of everything that this package is capable of, it is worth taking a look at the book The caret Package.
training$class <- as.factor(training$class)
# random forest
predictors = training[,3:9]
response = training[,"class"]
The first step here is to define your response and predictors. The response is simply one column that contains your class label, because we want to classify the rest of the image. The predictors are all of the columns with the information that we extracted from your raster stack. Be careful not to include anything else in your dataframe (e.g. the geometry or additional IDs).
Now, use the caret package to train a simple random forest model. We can use either the method “rf” or “ranger” in the train function to do so. There are many other implementations of the random forest algorithm in R that you can explore, for example the ranger package, which performs better.
Note
We also use the expand.grid function to do some model tuning, as well. While it seems complicated, this essentially returns a model with the parameters optimized for the best accuracy.
# create grid for tuning features
tgrid <- expand.grid(
mtry = 2:4,
splitrule = "gini",
min.node.size = c(10, 20)
)
The function returns a “tuning grid” that contains several possible combinations of parameters for example:
| mtry | splitrule | min.node.size | |
| 1 | 2 | gini | 10 |
| 2 | 3 | gini | 10 |
| 3 | 4 | gini | 10 |
| 4 | 2 | gini | 20 |
| 5 | 3 | gini | 20 |
| 6 | 4 | gini | 20 |
One model is trained for each of these six combinations of parameters. You can then choose the set of the parameters that performs best for your final model. But be careful if you are using this method – you are calculating not one but several models. The computation time will be a lot higher!
If you don’t want to tune your model set just one option for every parameter (e.g. mtry = 4).
You can speed up the model training by using parallelization (depending on the performance of your PC). We can use the detectCores() function, which automatically uses all available cores of the PC. To prevent your PC from bursting into flames, it is best to never use all cores (here we leave out two).
# create parallel cluster to increase computing speed
n_cores <- detectCores() - 2
cl <- makeCluster(n_cores)
registerDoParallel(cl)
# control the parameters of the train function
ctrl <- trainControl(method="cv",
number = 10, # number of folds
savePredictions = TRUE,
allowParallel = TRUE)
# train the model
model <- train(predictors,
response,
method = "ranger",
trControl = ctrl,
tuneGrid = tgrid,
num.trees = 100,
importance = "permutation")
# stop parallel cluster
stopCluster(cl)
# save the model
saveRDS(model, file.path(envrmt$path_models, "model.RDS"))
Congratulations, you have a fully developed random forest model! Now, you can use your model to classify the whole study area. Take a closer look at the accuracy and Kappa values as well as the variable importance for your model. What stands out to you about the values?
Prediction
Let’s admire our results now. Use your finished model to make a prediction on your predictor raster stack.
model <- readRDS(file.path(envrmt$path_models, "model.RDS"))
# use model to make a spatial prediction (SpatRaster)
prediction <- raster::predict(rasterStack, model, na.rm = TRUE)
# save prediction raster
raster::writeRaster(prediction, file.path(envrmt$path_prediction, "pred.tif"), overwrite = TRUE)
# save SpatRaster as RDS
saveRDS(prediction, file.path(envrmt$path_prediction, "pred.RDS"))
Your prediction may look something like the map below. As you can see, this model did not manage to distinguish the roofs of the houses from other sealed surfaces, such as roads and parking lots. Even some of the fallow fields were classified as buildings. But the results of your model may look quite different. In general, the quality of a model depends to a large degree on its training areas, so try to cover as wide of a spectrum as possible with your training classes.
Full screen version of the map
Validation
Finally, we need one more very important thing: Validation through independent data that was not included in the training of the model. For this, we use the data that we left out at the very beginning and that was not used to train the model. We predict values for each pixel and compare them with the ground truth values in a matrix.
test_data = readRDS(file.path(envrmt$path_model_training_data, "extr_test.RDS"))
mod = readRDS(file.path(envrmt$path_models, "model.RDS"))
predicted = stats::predict(object = mod, newdata = test_data)
val_df = data.frame(ID = dplyr::pull(test_data, "OBJ_ID"),
Observed = dplyr::pull(test_data, "class"),
Predicted = predicted)
val_cm = confusionMatrix(table(val_df[,2:3]))
# output
saveRDS(val_cm, file.path(envrmt$path_validation, "confusionmatrix.RDS"))
You have created a so-called confusion matrix. This gives you an initial impression about the model’s performance. The confusion matrix shows you which classes get misinterpreted for which other class.
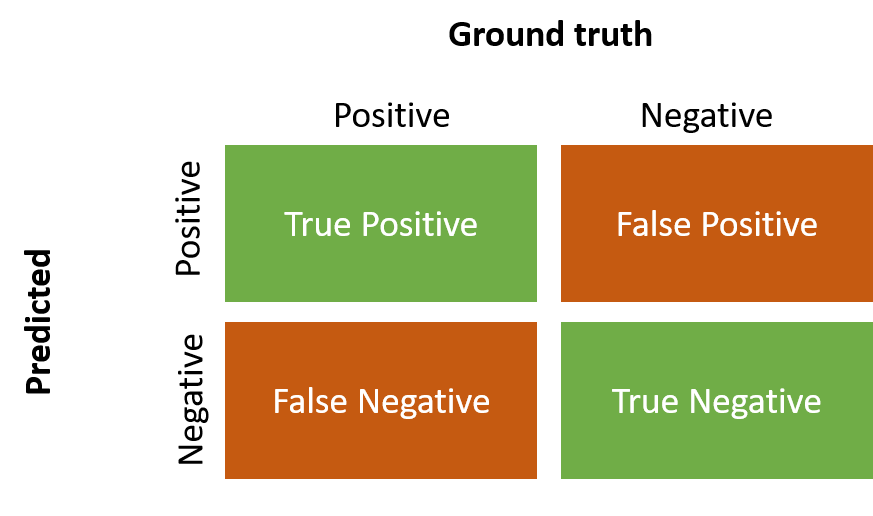
Image: Confusion Matrix
We will have a look at two common used performance metrics of the models now. The accuracy of the model shows the proportion of true positives and true negatives among all observations. If you have a more imbalanced dataset you can use Kappa, which gets calculated similar to accuracy but takes into account the possibility that an agreement happened due to chance, therefore it is more robust.
Comments?
You can leave comments under this Issue if you have questions or remarks about any of the code chunks that are not included as gist. Please copy the corresponding line into your comment to make it easier to answer the question.
