| EX | Create masks |
In this exercise, we will create a raster mask from vector data containing the outlines of all buildings in our study area in the southern part of Marburg (map below). For this purpose, both the raster mask and the DOP of the study area will be split from large raster (.tif) files into many smaller image files (.png).
Set up a working environment
First, set up the working environment as described in Unit 01. We will add some more packages and an additional folder to your setup script. In the “modelling” folder you can store your prepared data, your models, your predictions etc.
require(envimaR)
packagesToLoad = c(
"terra",
"png",
"tensorflow",
"keras",
"reticulate",
"sf",
"osmdata",
"rsample",
"tfdatasets",
"purrr",
"stars",
"magick"
)
# define a project rootfolder
rootDir = "~/edu/geoAI"
# some new paths
projectDirList = c(
"data/",
"data/modelling/",
"data/modelling/model_training_data/",
"data/modelling/model_training_data/dop/",
"data/modelling/model_training_data/bui/",
"data/modelling/models/",
"data/modelling/prediction/",
"data/modelling/validation/",
"data/modelling/model_testing_data/",
"data/modelling/model_testing_data/dop/",
"data/modelling/model_testing_data/bui/",
"docs/",
"run/",
"tmp",
"src/",
"src/functions/"
)
# Now set automatically root direcory, folder structure and load libraries
envrmt = envimaR::createEnvi(
root_folder = rootDir,
folders = projectDirList,
path_prefix = "path_",
libs = packagesToLoad,
alt_env_id = "COMPUTERNAME",
alt_env_value = "PCRZP",
alt_env_root_folder = "F:/BEN/edu"
)
## set terra temp path
terra::terra(tempdir = envrmt$path_tmp)
Read the data
Load the data and crop it to the extent of the Marburg DOP.
# read data
ras <- terra::rast(file.path(envrmt$path_data, "marburg_dop.tif"))
# subset to three channels
ras <- subset(ras, c("red", "green", "blue"))
# download and crop the OSM building data to the extent of the raster data
buildings = opq(bbox = "marburg de") %>%
add_osm_feature(key = "building") %>%
osmdata_sf()
buildings = buildings$osm_polygons
buildings = sf::st_transform(buildings, crs(ras))
ras_extent <- ext(ras)
buildings <- sf::st_crop(buildings[1], ras_extent)
Full screen version of the map
Rasterize the buildings
From the file shown in the figure above, containing the outlines of the buildings in the study area, we will first create a raster mask. To do this, we use the DOP of the study area as a reference raster, to ensure that the mask will have the same extent and the same resolution. The transformation of polygons into a raster file with the same properties as the DOP is done by the function rasterize of the raster package. Afterwards a reclassification of the values is performed, where all values that do not represent a building are 0 and all values that represent a building are 1. You can roughly see how the mask for the study area might look in the map below (layer mask).
# rasterize the buildings
rasterized_vector <- rasterize(buildings, ras[[1]])
# reclassify to 0 and 1
rasterized_vector[is.na(rasterized_vector[])] <- 0
rasterized_vector[rasterized_vector > 1] <- 1
#save
terra::writeRaster(rasterized_vector,
file.path(envrmt$path_data, "marburg_mask.tif"),
overwrite = T)
Full screen version of the map
Divide dataset to training and testing
Now we will cut the DOP and the mask in two pieces. You can use the extents from below, or choose two on your own from the image. The larger section will be used to train the model, while the smaller section will be used for testing.
# divide to training and testing extent
e_test <- extent(483000, 484000, 5626000, 5628000)
e_train <- extent(480000, 483000, 5626000, 5628000)
marburg_mask_train <- crop(rasterized_vector, e_train)
marburg_dop_train <- crop(ras, e_train)
marburg_mask_test <- crop(rasterized_vector, e_test)
marburg_dop_test <- crop(ras, e_test)
writeRaster(
marburg_mask_test,
file.path(envrmt$path_model_testing_data, "marburg_mask_test.tif"),
overwrite = T
)
writeRaster(
marburg_dop_test,
file.path(envrmt$path_model_testing_data, "marburg_dop_test.tif"),
overwrite = T
)
writeRaster(
marburg_mask_train,
file.path(envrmt$path_model_training_data, "marburg_mask_train.tif"),
overwrite = T
)
writeRaster(
marburg_dop_train,
file.path(envrmt$path_model_training_data, "marburg_dop_train.tif"),
overwrite = T
)
Function to split the data
To train the U-Net, many smaller images of the same size are needed instead of the large raster files we have at the moment. We will use the function below to split the raster files. Although it may seem a bit complex at first glance, it basically consists of five simple steps:
- determine the size of the original raster (DOP)
- determine how many images of a certain size (e.g. 128 by 128) can fit into it and how large the extent of all the images is in total.
- crop the original grid to the new size (a multiple of 128x128).
- create a grid with polygons over the cropped raster (see layer images in the image below)
- crop every polygon to both the house mask and the DOP and save the small 128x128 images as .png
subset_ds <- function(
input_raster,
model_input_shape,
path,
targetname = ""
) {
targetSizeX <- model_input_shape[1]
targetSizeY <- model_input_shape[2]
inputX <- ncol(input_raster)
inputY <- nrow(input_raster)
diffX <- inputX %% targetSizeX
diffY <- inputY %% targetSizeY
# determine new dimensions of raster and crop,
# cutting evenly on all sides if possible
newXmin <- ext(input_raster)[1] + ceiling(diffX / 2) * res(input_raster)[1]
newXmax <- ext(input_raster)[2] - floor(diffX / 2) * res(input_raster)[1]
newYmin <- ext(input_raster)[3] + ceiling(diffY / 2) * res(input_raster)[2]
newYmax <- ext(input_raster)[4] - floor(diffY / 2) * res(input_raster)[2]
rst_cropped <- crop(
input_raster,
ext(newXmin, newXmax, newYmin, newYmax)
)
# grid for splitting
agg <- terra::aggregate(
rst_cropped[[1]],
c(targetSizeX, targetSizeY)
)
agg[] <- 1:ncell(agg)
agg_poly <- as.polygons(agg)
names(agg_poly) <- "polis"
# split and save
lapply(seq_along(agg), FUN = function(i) {
subs <- local({
e1 <- ext(agg_poly[agg_poly$polis == i, ])
subs <- crop(rst_cropped, e1)
})
writePNG(
as.array(subs),
target = file.path(path, paste0(targetname, i, ".png"))
)
})
# free memory
rm(agg, agg_poly)
gc()
return(rst_cropped)
}
Full screen version of the map
Function to remove files without training data
In this example we will only use the images to train the U-Net that also contain one of the objects we want to detect (a building). Therefore we will use the following function to remove from all .pngs, that have only one value in the mask (0 no house), both the mask image and the corresponding DOP .png.
# remove all masks with just background information and
# keep the foreground (building) information
remove_files <- function(df) {
lapply(seq(1, nrow(df)), function(i) {
local({
fil = df$list_masks[i]
png = readPNG(fil)
len = length(png)
if((sum(png) == len) | (sum(png) == 0)) {
file.remove(df$list_dops[i])
file.remove(df$list_masks[i])
}
})
})
return("Done")
}
Subset the datasets
Both the DOP and the mask are split into smaller .pngs using the subset_ds function. The output path for both functions is in a different folder, where the images should be stored.
# read training data
marburg_mask_train <-
stack(file.path(envrmt$path_model_training_data, "marburg_mask_train.tif"))
marburg_dop_train <-
stack(file.path(envrmt$path_model_training_data, "marburg_dop_train.tif"))
# set the size of each image
model_input_shape = c(128, 128)
# subsets for the mask
subset_ds(
input_raster = marburg_mask_train,
model_input_shape = model_input_shape,
path = envrmt$path_model_training_data_bui
)
# subsets for the dop
subset_ds(
input_raster = marburg_dop_train,
model_input_shape = model_input_shape,
path = envrmt$path_model_training_data_dop
)
Remove empty files
Now we can apply our functions defined above to our data. Both the DOP and the mask are split into smaller .pngs using the subset_ds function. The output path for both functions is in a different folder, where the images should be stored.
# list all created files in both folders
list_dops <-
list.files(envrmt$path_model_training_data_dop,
full.names = TRUE,
pattern = "*.png")
list_masks <-
list.files(envrmt$path_model_training_data_bui,
full.names = TRUE,
pattern = "*.png")
# create a data frame
df = data.frame(list_dops, list_masks)
remove_files(df)
Expected output
At the end of this exercise you should have created a raster mask from the vector file and created a subset of the DOP and the mask which you can finde in two different folders, the images should look similar to the images below.
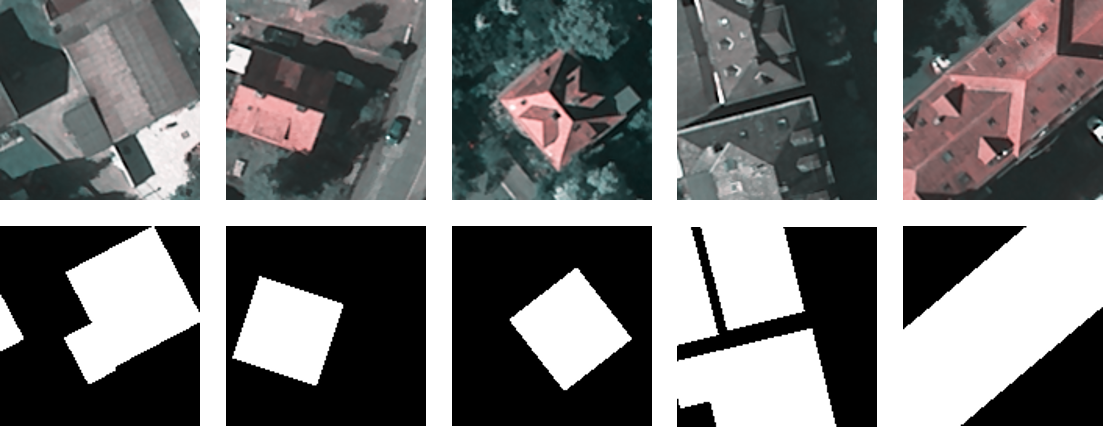 Image: © OpenStreetMap contributors; Hessische Verwaltung für Bodenmanagement und Geoinformation
Image: © OpenStreetMap contributors; Hessische Verwaltung für Bodenmanagement und Geoinformation
Comments?
You can leave comments under this Issue if you have questions or remarks about any of the code chunks that are not included as gist. Please copy the corresponding line into your comment to make it easier to answer the question.
